RootScape
Coruzzi lab @ New York University
Overview
| Measured variables | shape |
|---|---|
| Operating system | windows, mac, linux |
| Licence | freeware, matlab |
| Automation level | semi-automated |
| Plant requirements | arabidopsis |
| Export formats | csv |
| Other information | - |
Scientific article(s)
RootScape: A Landmark-Based System for Rapid Screening of Root Architecture in ArabidopsisD. Ristova,U. Rosas,G. Krouk,S. Ruffel,K. D. Birnbaum,G. M. CoruzziPLANT PHYSIOLOGY, 2013 View paper
Similar tools
Other tools for the analysis of root-system:
Description
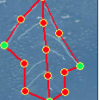
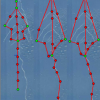
RootScape is a landmark-based allometric method for rapid phenotyping of Root System Architecture. Using the software AAMToolbox, we created a 20- point landmark model that captures RSA as one integrated trait, and used this model to quantify changes in the RSA of Arabidopsis (Col-0) wild-type plants grown under different hormone treatments. Principal Component Analysis (PCA) was used to compare RootScape to conventional methods designed to measure root architecture. This analysis showed that RootScape efficiently captured nearly all the variation in root architecture detected by measuring individual root traits, and is five-to-ten times faster than conventional scoring. RootScape was validated by quantifying the plasticity of RSA in several mutant lines affected in hormone signaling. The RootScape analysis recapitulated previous results that described complex phenotypes in the mutants and identified novel gene-by-environment interactions.
Source: Ristovaet al. (2013) Plant Physiology , 161(3), 1086-96