MARS-ALT
Virtual Plant @ INRIA
Overview
| Measured variables | n-cells, volume, shape, 3D-reconstruction, surface |
|---|---|
| Operating system | windows, mac, linux |
| Licence | freeware |
| Automation level | semi-automated |
| Plant requirements | any |
| Export formats | open alea |
| Other information | Works as a module of OpenALea |
Scientific article(s)
Imaging plant growth in 4D: robust tissue reconstruction and lineaging at cell resolutionRomain Fernandez,Pradeep Das,Vincent Mirabet,Eric Moscardi,Jan Traas,Jean-Luc Verdeil,Grégoire Malandain,Christophe GodinNature Methods, 2010 View paper
Similar tools
Other tools for the analysis of cell:
Description
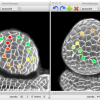
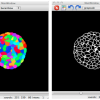
Quantitative information on growing organs is required to better understand morphogenesis in both plants and animals. However, detailed analyses of growth patterns at cellular resolution have remained elusive. We developed an approach, multiangle image acquisition, three-dimensional reconstruction and cell segmentation-automated lineage tracking (MARS-ALT), in which we imaged whole organs from multiple angles, computationally merged and segmented these images to provide accurate cell identification in three dimensions and automatically tracked cell lineages through multiple rounds of cell division during development. These methods were used to quantitatively analyze Arabidopsis thaliana flower development at cell resolution, which revealed differential growth patterns of key regions during early stages of floral morphogenesis.
Source: Fernandezet al. (2010) Nature Methods , 7(7), 547-53