RootScan
Roots lab @ Penn State University
Overview
| Measured variables | anatomy |
|---|---|
| Operating system | windows, mac, linux, matlab |
| Licence | freeware |
| Automation level | automated |
| Plant requirements | any, radial section |
| Export formats | - |
| Other information | - |
Scientific article(s)
RootScan: Software for high-throughput analysis of root anatomical traitsAmy L. Burton,Michael Williams,Jonathan P. Lynch,Kathleen M. BrownPlant and Soil, 2012 View paper
Similar tools
Other tools for the analysis of single-root:
Description
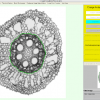
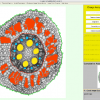
RootScan is a program for automated image analysis of anatomical traits in root cross-sections. RootScan uses pixel thresholds to separate the cross-section from its background and to divide it into tissue regions. Area measurements and object counts are performed within various regions of interest. A graphical user interface permits the user to see which regions are selected, to edit those selections, and to rate and comment on the data. The structure of†the program allows for organized workflow and in- creased data collection efficiency. The program collects data on more than 20 variables per image including areas of the cross- section, stele, cortex, aerenchyma lacunae, xylem vessels, and counts of cortical cells and cell files. An increased rate of data collection allows collection of four times more variables in less time than is possible with current methods. Correlation analysis shows that RootScan data is equal or greater in accuracy than data collected with Photoshop.
Source: Burton et al (2012) Plant and Soil, 357(1-2), 189-203