rhizoTrak
Martin Luther University Halle-Wittenberg and Leipzig University
Overview
| Measured variables | length, diameter, topology |
|---|---|
| Operating system | windows, mac, linux |
| Licence | open-source, freeware |
| Automation level | manual |
| Plant requirements | any |
| Export formats | rsml, csv |
| Other information | Works as a FIJI plugin |
Scientific article(s)
rhizoTrak: A flexible open source Fiji plugin for user-friendly manual annotation of time-series images from minirhizotronsBirgit Moeller,Hongmei Chen,Tino Schmidt,Axel Zieschank,Roman Patzak,Manfred Tuerke,Alexandra Weigelt,Stefan PoschbiorXiv, 2019 View paper
Similar tools
Other tools for the analysis of root-system:
Description
rhizoTrak is:
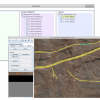
- an open source tool for flexible and efficient manual annotation of complex time-series minirhizotron images
- building on TrakEM2,
- realized as a Fiji plugin available for Windows, Linux, and MacOS.
- import and processing of the complete set of images of a time series in one single project
- easy browsing through time points which enables to annotate roots by simultaneously considering images of different time points
- temporal links between roots in different images which explicitly represented identity of the same physical root via so-called connectors
- easy transfer of annotation data of one time point to the next as initial annotation where connectors between corresponding root annotations are automatically established
- automatic checks for inconsistencies in root typology due to merging of roots and assistance in solving them
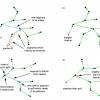
- the inherent representation of branching roots by modeling roots as treelines
- an individual radius for each node of a treeline, and user-definable status labels for segments of treelines
- a large collection of editing operations for root treelines including split and merge, where many of these operations are easily accessible via keyboard shortcuts
- different visualization options for treelines and fast hiding and un-hiding of all annotated treelines
- redirection of treelines which is important as the upstream and downstream of a root is often not straightforward to be identified in minirhizotron images
- import and export of annotations from/to RSML format,
- export of root measurements in a tab-separated format including the option to aggregate measurements within images
- a plugin mechanism to merge results from automatic segmentation method which are to be implemented as a MiToBo operator